From radioactive sea monsters to mutant superbugs. This week, I’ll be discussing antibiotic-resistant viruses and their relationship with wastewater treatment!
Antibiotic compounds have flooded and permeated into every nook and cranny of our society. They are commercially utilized for livestock and aquaculture to prevent diseases outbreaks; applied to crops to improve disease resistance and increase yields; and even prescribed by clinics and hospitals(Kümmerer, 2009) for minor ailments like giving candy to children during Halloween!
Antibiotic introduction into the natural environment come from both non-point sources (surface run-off, groundwater permeation etc.) and this post’s primary focus: point source from wastewater treatment plants(WWTP) and direct discharge(Kümmerer, 2009). However, the real danger does not arise from the antibiotics released itself, but what happens when bacteria and viruses present and utilized in biochemical treatment in WWTPs receive a sublethal dose of antibiotics(Manaia et al., 2018).
That’s right! For the same reason your doctor tells you to finish the prescribed dose of antibiotics, a non-lethal dose of antibiotics combined with bioreactor conditions that boost bacterial growth hosts a favourable environment for bacteria with antibiotic-resistant genes(ARG) to outcompete their regular counterparts(Manaia et al., 2018).
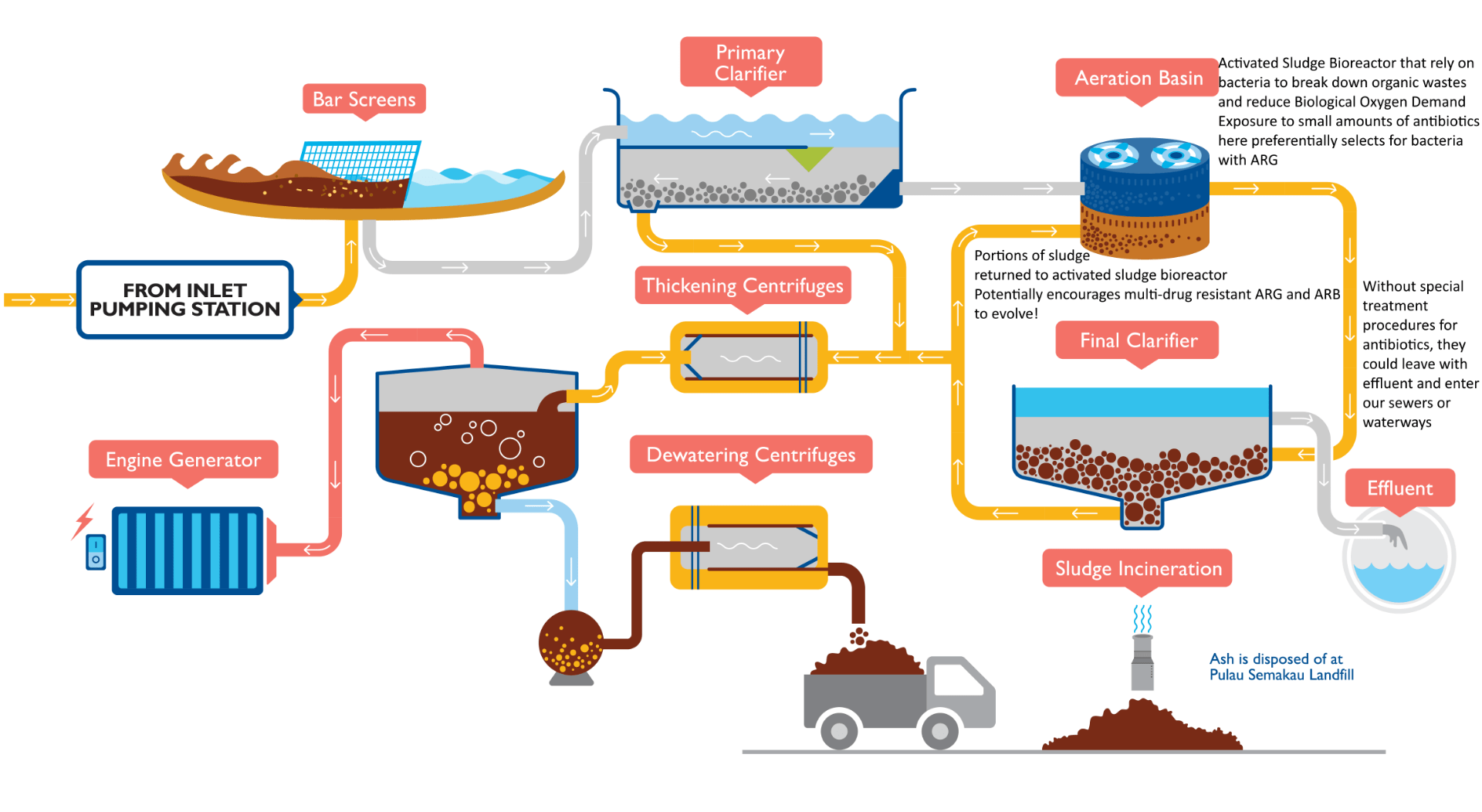
Potential for ARG and ARB to develop in PUB’s wastewater treatment system. Source: https://www.pub.gov.sg/PublishingImages/PUB_29_UsedWaterConventionalTreatment%20PA-01.png
To make things worse, many bioreactors (like in PUB’s municipal WWTPs) recycles portions of bacterial sludge from previous batches, exposing the antibiotic-resistant bacteria(ARB) to a wide variety of other antibiotics(Manaia et al., 2018). These fluctuating conditions preferentially select bacteria and viruses with ARG that resist a wide variety of antibiotics(Manaia et al., 2018), similar to Dr Rick Pott’s Variability Selection hypothesis.
If you’ve read A Clearer Picture, you might be wondering: since the wastewater ends up being disinfected before being discharged, why worry? While most ARB are vulnerable to disinfection, some studies show that ARG go through WWTPs largely untouched where they may be transferred to other cells in the natural environment(Yuan, Guo, & Yang, 2015); some ARB can even go into dormancy only to be reactivated once out of the disinfection tank(Manaia et al., 2018). Of course, antibiotics also make it out of the treatment process(Kümmerer, 2009), which makes it easier for bacteria in natural ecosystems to evolve into ARBs.
The consequences of this inadequacy are all around us. Wei et al. (2018) found significant amounts of ARG in bioreactor bacteria in Chinese WWTPs; Hatosy & Martiny (2015) found known and undiscovered ARG in coastal waters of California and Hawaii; and to top it all off, two separate studies found ARBs in bottlenose dolphins, harbour seals and harbour porpoises ((Schaefer et al., 2019), and this preliminary study conducted in Puget Sound).

A Harbour Porpoise, one of the hosts of ARB found in a preliminary study. Harbour Porpoise ©Niki Clear. Source: https://www.wildlifetrusts.org/wildlife-explorer/marine/marine-mammals-and-sea-turtles/harbour-porpoise
All that is left for a potentially apocalyptic outbreak is for the ARG to incorporate itself in a human vector or pathogen(like E. Coli) or god forbid, an influenza virus with multiple ARGs that resists all kinds of antibiotics, a superbug(Manaia, 2017).

Possible Pathways for ARG and ARB to make their way from natural systems into human and livestock. Source: (Manaia et al., 2017)
If it isn’t clear enough already, excessive antibiotic use not the way forward. With ARG all over our biosphere, it’s just a matter of time before a superbug pandemic matching the scale of the black plague starts. Antibiotic substitutes like Yan Yang et al.’s polymer could be our salvation. Until then, go organic, eat fewer products that use antibiotics(like meat and dairy); and please, finish your antibiotic dose. Doctor’s orders!
Next week, I’ll be looking at the ways to deal with antibiotics and pharmaceuticals right at the source, so stay tuned!
References:
Hatosy, S. M., & Martiny, A. C. (2015). The ocean as a global reservoir of antibiotic resistance genes. Applied and Environmental Microbiology. https://doi.org/10.1128/AEM.00736-15
Kümmerer, K. (2009). Antibiotics in the aquatic environment – A review – Part I. Chemosphere. https://doi.org/10.1016/j.chemosphere.2008.11.086
Manaia, C. M. (2017). Assessing the Risk of Antibiotic Resistance Transmission from the Environment to Humans: Non-Direct Proportionality between Abundance and Risk. Trends in Microbiology. https://doi.org/10.1016/j.tim.2016.11.014
Manaia, C. M., Rocha, J., Scaccia, N., Marano, R., Radu, E., Biancullo, F., … Nunes, O. C. (2018). Antibiotic resistance in wastewater treatment plants: Tackling the black box. Environment International. https://doi.org/10.1016/j.envint.2018.03.044
Schaefer, A. M., Bossart, G. D., Harrington, T., Fair, P. A., McCarthy, P. J., & Reif, J. S. (2019). Temporal Changes in Antibiotic Resistance Among Bacteria Isolated from Common Bottlenose Dolphins (Tursiops truncatus) in the Indian River Lagoon, Florida, 2003-2015. Aquatic Mammals, 45(5), 533–542. https://doi.org/10.1578/AM.45.5.2019.533
Wei, Z., Feng, K., Li, S., Zhang, Y., Chen, H., Yin, H., … Deng, Y. (2018). Exploring abundance, diversity and variation of a widespread antibiotic resistance gene in wastewater treatment plants. Environment International. https://doi.org/10.1016/j.envint.2018.05.009
Yuan, Q. Bin, Guo, M. T., & Yang, J. (2015). Fate of antibiotic resistant bacteria and genes during wastewater chlorination: Implication for antibiotic resistance control. PLoS ONE. https://doi.org/10.1371/journal.pone.0119403